This story was originally published by Ken Stranderg with Scientific Computing.

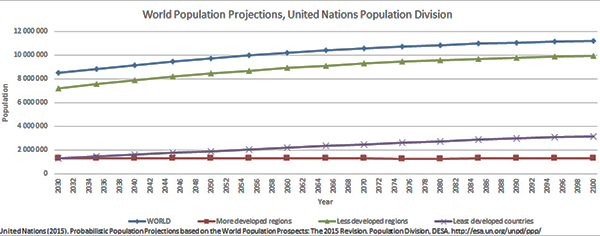
According to the United Nations, the world’s population will be over 9 billion by 2050 and over 11 billion by the next century. That’s a lot of souls for which the world’s farmers — both agriculturists and aquaculturists — must produce nutrient-rich foods.
In 2009, the United Nations Food and Agriculture Organization (FAO) held a High Level Experts Forum in Rome to discuss food needs around the planet. The Forum projected that feeding over 9 billion people in 2050 would “require raising overall food production by some 70 percent between 2005/07 and 2050. Production in the developing countries would need to almost double.”[1]
Research from the University of Minnesota[2]indicates that maize, rice, wheat and soybean crop yields are not on track to meet the food demand of the world’s population in 2050, and many experts agree that, rather than turning more rain forests into farmland, agriculture must instead focus on increasing crop yields.
ISU on the forefront of food science
Increasing the quality and yield of food crops and animals has been a focus of agriculture for many years, and Iowa State University is well-known for its contributions to agriculture and other biological sciences. “ISU has large programs in agricultural science, veterinary medicine, agricultural economics, land use and climate modeling,” said Arun Somani, Associate Dean for Research in the university’s College of Engineering. “We’ve been bringing together researchers in climate, agriculture, biology and bioinformatics to solve problems around cropping in changing climates, how we produce food, and how to help the industry improve production.” This is no surprise, considering Iowa is at the forefront of food-producing states in America. Iowa is among the largest producer of corn[3] and pork,[4] and among the top 10 for cattle.[5]
Improving corn through the ancestry of maize
One of those researchers is Matthew Hufford, Assistant Professor of Ecology, Evolution and Organismal Biology. “Maize, or corn, is one of the highest producing crops in the world and is very important to the Iowan economy,” said Dr. Hufford. His research specializes in teosinte, the wild ancestor of corn, and evolution of the domesticated maize we grow today. He looks for interesting genomic differences that may have led to an improvement in the quality of corn and the ability to grow it more effectively.
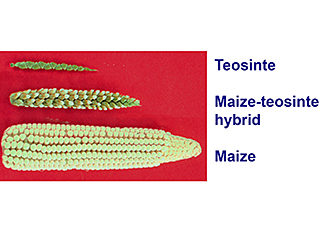
“We believe that corn — Zea mays subspecies mays — was domesticated from teosinte, which is a collective term to refer to wild species in the genus Zea,” explained Hufford. “The closest wild relative to corn is Zea mays subspecies parviglumis, found only in southwest Mexico. Additional teosinte subspecies and species are found throughout Mexico and Central America.”
Hufford looks for interesting adaptations across teosinte taxa that might lend themselves to improvements if bred into domesticated corn. For example, another teosinte, Zea luxurians, which is found in southeast Guatemala, forms hollow tube structures called aerenchyma that go through the entire stem. These structures are common in wetland plants.

Waterlogged soils become hypoxic (low in oxygen) when other organisms consume the soil’s oxygen before the plant can. The aerenchyma bring oxygen down to the roots. With the increasing rains in the Midwest over the last few decades, the soils carry much more standing water than in the past (see article), which has affected the health of crops, such as soybeans, and delayed or prevented plantings of other crops, such as corn. Breeding such structures into corn might have a valuable impact on their ability to grow better in the wetter soils of today’s Midwest.
Professor Hufford said that most of the work done in the Hufford Lab deals with genomic data. “We look at two different time points in terms of the evolutionary past of maize. We collect individuals of teosinte in the regions where they still grow, and then sequence their genomes. Then we study each genome and look for really marked differences, genome-wide, that could underlie the whole process of domestication. When we find regions of the ancient genome that look interesting, we annotate them, and the plant breeders can take this information and potentially improve the genotype of maize.”
That’s the first evolutionary comparison Hufford makes. He’s also interested in how maize has spread across the landscape to different regions since it was domesticated some 9,000 years ago. Maize cultivation began in a very confined region of southwest Mexico, but by the time Columbus came to the Americas, maize grew all the way from the Andes up to Canada. “So, it spread and adapted to all these unique conditions involving drought tolerance, flood tolerance, adaptation to high elevations, and more,” stated Hufford.
Hufford and his team collect samples of ancient varieties of maize, called landraces, from across the regions, and they sequence their genomes. Then they compare the genomes of landraces with an interesting adaptation (e.g., the ability to grow at high elevation) to genomes of maize lacking this adaptation. “We look for differences that might indicate why a particular landrace grows so well in a given environment,” said Hufford. “When we publish these adaptation loci, the entire study becomes part of the public domain for interested parties, such as breeders and other researchers.”
In Professor Hufford’s work, each maize genome is around 3 gigabases (which equals about 3 Gbytes). But to reliably get information from the genome, he has to generate data as many as 150 times the size of the genome. That means they end up processing a lot of data.
Genome informatics @ ISU
Many researchers at Iowa State, such as Professor Hufford, work in collaboration with the Genome Informatics Facility (GIF), a part of the Office of Biotechnology at ISU. The facility was established in 2011 to enable scientific discovery through the transformation of big data into data that dramatically accelerates our understanding of biology and evolutionary processes. It relies on the high-performance computing clusters at ISU, utilizes open access/open source software, and is managed by Dr. Andrew Severin.
“Genome sequencing is a challenging problem,” commented Severin. “The data generated for genomics are expected to be greater than many other big data fields in the near future, including astronomy, Twitter and YouTube.[6] There’s that much data. And, the amount of genomic data that can be acquired roughly quadruples every year.”
Assembling a genome is a very large, compute-intensive task that begins with sequencing. Gene sequencing machines create data sets, called reads, of DNA base sequences, comprising a few hundred to a few thousand bases. There is overlap, where the ends of some reads are the same as the ends of other reads, creating a clue for how the sequences join together to create the genome. “Assembly is like putting together a gigantic jigsaw puzzle of millions and millions, perhaps billions, of pieces,” stated Severin. “The computer compares every sequence to every other sequence to see if the two pieces fit together and then begins to assemble them piece by piece. It’s a very serial process.” Assembly is further complicated, because a large portion of the DNA has 30 percent or more of pieces that have identical overlaps, yet appear in different positions across the DNA molecule. In addition, current sequencing technology can introduce pieces with errors in them that need to be accounted for in order for the assembly to come together well. It takes massive amounts of processing.
“Within the Facility, we average over 100,000 CPU hours a month,” stated Severin. “That is equivalent to 69 duo-core laptops running simultaneously for every hour of every day for 30 days.”
Condo (short for Condominium) is the latest HPC cluster at ISU, and it represents a new way that ISU supports HPC. Instead of the traditional method of each department/research group buying and managing its own system, a large shared machine is acquired and managed centrally. Each department/research group contributes to purchases of only the computing nodes and storage (the computing resources) to meet its average need. All users share the computing resources up to their allocation on average, or more in case some unused cycle are available “This way, there is plenty of computing for any researcher that needs to run a really big job,” said Somani. “Now, it’s the way HPC is done at ISU.”
Condo houses 156 main nodes (expandable to 324), each with two 8-core Intel processors, 128 GB of memory, and 2.5 TB of local storage. Additionally, Condo has two large memory nodes: one with 40 Intel processor cores and 2 TB of memory — purchased by Severin’s department specifically for genome assemblies — and one 32-core, 1 TB node. The system uses Intel True Scale Fabric architecture and the Intel Enterprise Edition for Lustre software parallel file system for high-performance storage.
“Until Condo, we ran much of our work on the HPC systems with a tenth of the power, such as Lightning 3, which are no longer sufficient,” stated Severin. “We prefer the Condo cluster, because it can facilitate trivial parallelization of common tasks, increasing our efficiency, and the large memory node is great for assembly.”
“Assemblies of genomes like teosinte require a lot of memory,” explained Hufford. “Some genomes we work with comprise 1.5 TB of raw data, uncompressed, which is about 1.7 billion reads. The computer essentially assembles the genome serially, so it must contain the entire set of reads in its memory during assembly. The process doesn’t lend itself to easy parallelization.”
“We first tried to run our assembly on Lightning 3,” continues Hufford, “but it ran out of memory really quickly. Then, we got access to the CyEnce cluster (a National Science Foundation funded project through Major Research Instrumentation and CISE Research Infrastructure grants) and used their 1 TB, high-memory node. We maxed that out, too.” Hufford and Severin eventually worked with the College of Liberal Arts and Sciences (LAS) IT department to build a 1.5 TB machine called Big RAM to complete his teosinte assembly in the interim before the Condo cluster became available.
Arun Seetharam assists Hufford with the programmatic side of his research. “I’m a biologist turned computer programmer,” stated Dr. Seetharam. “Big RAM was tuned for speed. It uses very high clock rate Intel processors with Turbo Boost, all solid-state drives (SSDs), and 1.5 TB of memory to hold all the reads. It’s extremely fast and suited for assembly. The pipeline of programs that I use is a mosaic of code derived from various open source programs. I tested a number of assemblers, such as the Discovar-denovo, MaSuRCA or ALLPATHS-LG, plus other programs, and picked the one that consistently performed the best on our dataset.”
“Once the genome is assembled,” adds Hufford, “we do a number of analyses, such as calculating a statistic around every gene in the genome. That process lends itself much more to parallelization.” Some components in Seetharam’s pipeline are trivially parallelizable, which he dispatches across all Intel cores spanning multiple nodes, but some are non-parallelizable and must execute serially. “Overall, I’d say 75 percent is parallelized,” commented Seetharam, “and 25 percent — the assembly — is serial, but the 25 percent takes 90 percent of the time.” As Seetharam ports and compiles his pipeline for Condo, which uses the Intel Compiler, he expects to run future genome assemblies on the 2TB Condo node, purchased specifically for that purpose.
Improving aquaculture through genomics
In addition to running the Genome Informatics Facility, Severin is working on projects involving sustainable domestic aquaculture. “Aquaculture today is where agriculture was about five to 10 years ago, in terms of genomic resources for selective breeding,” said Severin. “And, Iowa State is well-known for the agriculture and livestock genomes we’ve assembled and annotated. So, we’re applying the even-better sequencing technology that we have today with our genome assembly and annotation experience to aquaculture species.”
In collaboration with NOAA scientists, John Hyde and Catherine Purcell at the Southwest Fisheries Science Center, and Mark Drawbridge at Hubbs-SeaWorld Research Institute (HSWRI), Severin and GIF are developing genomic resources for Seriola dorsalis to improve the brood stock selection of Yellowtail Kingfish for grow-out. “There is so much we don’t know about an organism that can easily be addressed with sequencing data. For example, Yellowtail males and females are indistinguishable and can only be non-destructively sexed when they start releasing eggs or sperm. With the genomic resources we are developing, we hope to identify an area in the genome that can be used in a cost-effective, non-destructive (to the animal) gender test, permitting better ratios of male to female in the tanks and leading to increased production.”
Severin and collaborators were recently funded to expand the genomic resource development to other aquaculture species that include Albacore tuna, Seriola rivoliana, and five species of abalone (red, green, pink, white and black). Severin will take advantage of having genomes of multiple species for comparative genomic analyses, which he believes will bring an even deeper understanding of these organisms and how these organisms have evolved to thrive in their local environments.
Severin considers himself a tools expert. “A lot of what I do is create pipelines that can be used for genomic resources in the Facility to address a particular problem a biologist wants to solve,” he said. “These resources can then be used to improve the production of crops, livestock or aquaculture. We’ve run our own annotation pipeline on Condo and used Condo to parallelize what’s called our Maker-P pipeline. Condo will facilitate a whole new level of analysis that hasn’t been possible so far at ISU.”
Severin and his colleagues have been developing scripts to parallelize as many of their pipelines as possible for Seriola. One of the projects, he explains, is to run a very large transcriptome of 200,000 transcripts or gene products (a transcriptome is to RNA as a genome is to DNA) and parallelize it across 512 cores to identify other sequences in the transcriptome that are similar to the sequence that’s being worked on, and its annotation, or transcriptomes of its closest species.”
Severin has been able to run such projects across 512 Condo cores, reducing a job that would normally take more than 48-hours to just a few minutes. “Even if it’s something trivial, like formatting a file a particular way, it can take a really long time, running a single command serially on a file that’s several gigabases in size. On Condo, I can split the file into smaller pieces and run them in parallel across as many processors as there are available, trivially parallelizing the problem.”
HPC@ISU contributes to solving the world’s food challenges
According to the U.N., food shortages will become a serious problem for the world if we don’t improve how we produce our raw food supplies. Scientists at ISU, like Drs. Hufford, Seetharam and Severin, are part of a community solving problems in agriculture and aquaculture to help farmers grow crops more effectively. They’re using the most modern technologies and HPC computing clusters available to science today, contributing new areas of knowledge to science and farming.
“We get to see a lot of interesting biological problems,” said Severin. “Applying bioinformatic tools to help find solutions to those problems is very satisfying and a lot of fun!”
For further information:
- Genome Informatics Facility at ISU: http://gif.biotech.iastate.edu/
- Hufford Lab at ISU: http://www.public.iastate.edu/~mhufford/HuffordLab/home.html
- High-Performance Computing resources at ISU: http://www.hpc.iastate.edu/
- For a look at how a maize genome is depicted, see the Maize genome database browser: http://beta.maizegdb.org/gbrowse
Ken Strandberg is a technical story teller for technology areas that include software, HPC, industrial technologies, design automation, networking, medical technologies, semiconductor and telecom.
References
[3] U.S.D.A., “Crop Production 2014 Summary, January 2015,” http://www.usda.gov/nass/PUBS/TODAYRPT/cropan15.pdf